The brain; it takes one to know one
While you are reading this text, your brain is reading too. By making many complex computations and integrating information from many different brain regions, it ensures that you can make sense of the small dots, stripes and circles, that your eyes see. As a matter of fact, in the majority of our daily activities – walking, talking, playing a musical instrument – our brain is involved. It is the control station of our body, and yet we know so little about it. This is not surprising when you know that the brain contains about 86 billion brain cells, or neurons, and about 100 trillion connections. Finding out where in our brain all these daily activities are represented is looking for a needle in a haystack. It becomes even more complicated in the case of brain diseases such as dementia, Alzheimer’s disease, Parkinson’s disease or obsessive-compulsive disorder, where the brain is not fulfilling its important task correctly.
If we ever want to cure or prevent brain diseases, we need to build up a better understanding of how the brain works and how the disease evolves from a healthy brain. And for that, we need better tools. A big part of the problem is the brain’s complexity. We need a device that measures the activity of a multitude of neurons across regions, while maintaining a single-cell resolution. Until recently, these recording devices only succeeded in one or the other.
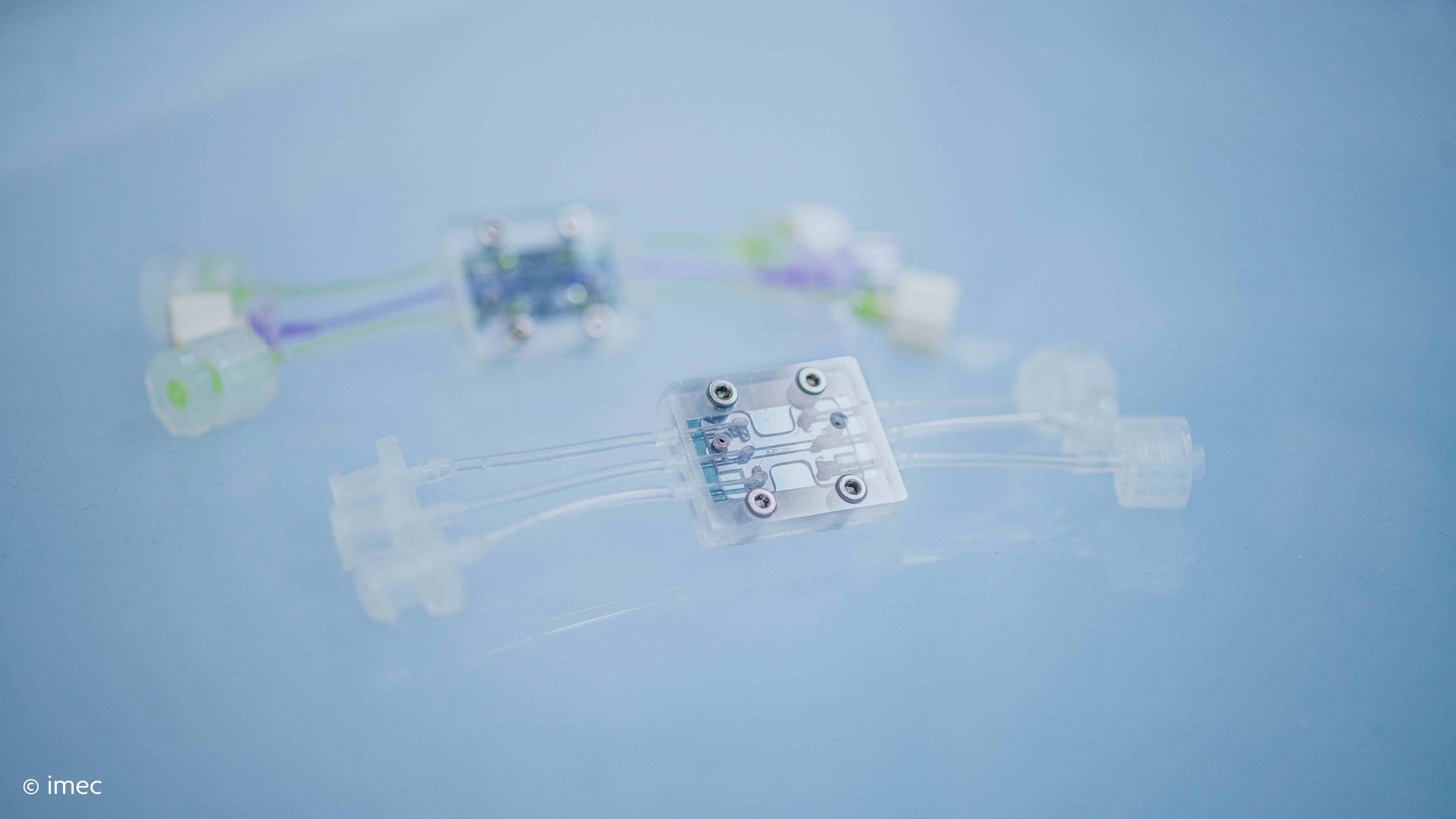
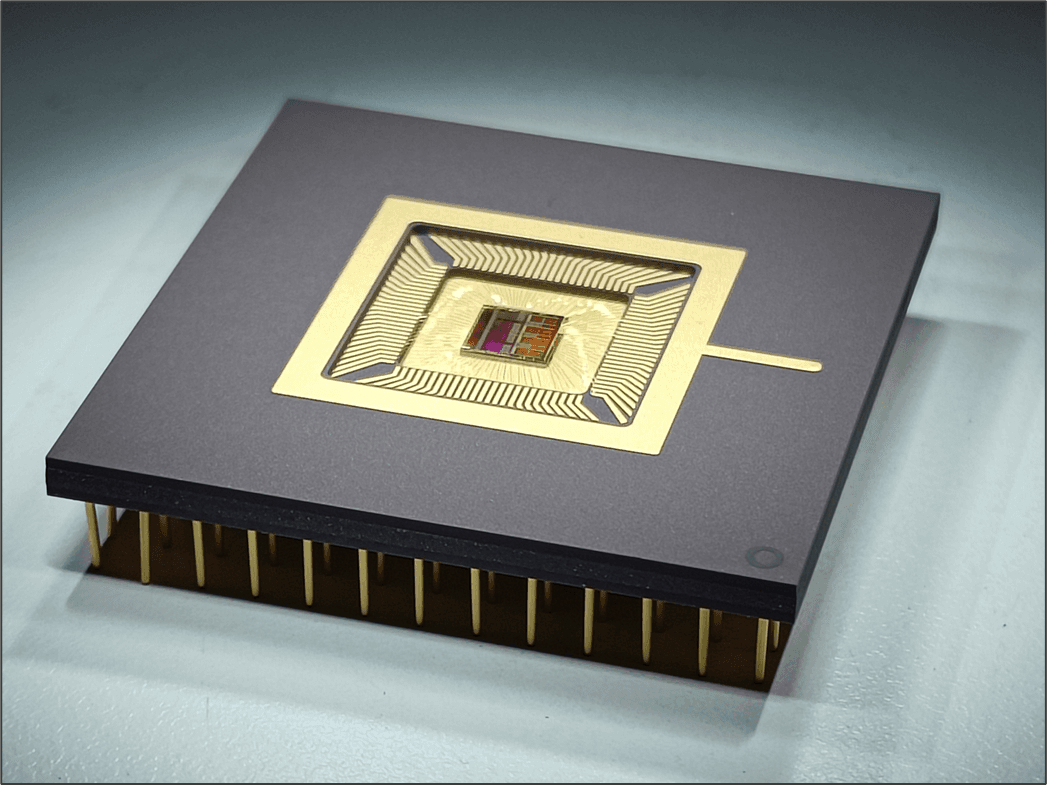
Now, imec has handed the brain researchers a new tool, the Neuropixels probe, that can interrogate 100s of neurons and many different brain regions simultaneously in a rodent bran, an order of magnitude more than current tools offer. With this ultra-thin, implantable, needle-like device, research groups have continued their mission to decipher the brain and decode the mechanisms behind brain disorders.
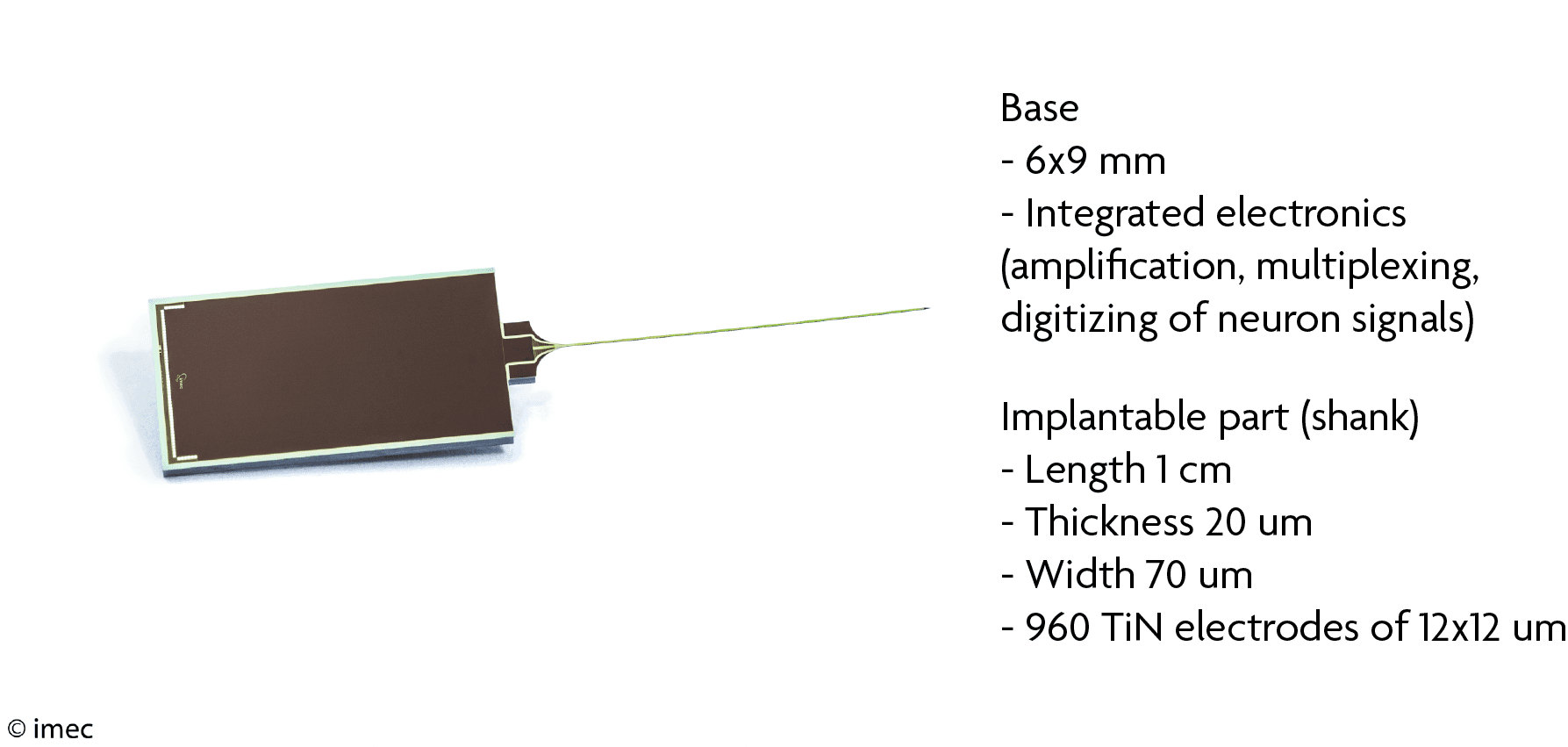
Neuropixels specifications
How does the brain make decisions?
We are confronted with decisions every day. Based on something we experience, how do we decide which action to pick among a selection of possible actions you can take, or rather how does your brain? Nick Steinmetz, senior scientist in the labs of Matteo Carandini and Kenneth Harris from University College London, took on the challenge to identify where decisions are formed in the brain. “Correlates of decision making have been described in many regions and systems of the brain and knowing where to start looking is not an easy task. One of the reasons for this is the strong interconnectedness of all of these structures that mixes up the incoming signals. Any signal that enters into the brain can in principle end up almost anywhere else in the brain, making it impossible to predict a priori where in the brain you might observe a particular signal. On the other hand, recurrent loops within and between these structures, coupled with non-linear features of the neurons, give rise to unpredictable dynamics, so that it is also impossible to predict a priori when a particular signal or process is going to take place in the brain.
In order to answer these very simple questions – when, where and how is a selection made between possible actions based on sensory information – you will need to measure these events. But, we need to measure at the level of single neurons and at millisecond time precision because that is the timescale of these sorts of behaviors.” The Neuropixels probes enable this kind of extensive and high-density recordings. They make it possible to sample the signals of single neurons from multiple brain regions simultaneously and continuously. Moreover, because the probe is so thin, you can fit multiple devices in the brain and cover more structures. That is exactly what Nick Steinmetz did, but in the context of a decision-making task in mice. He recorded the signals from 20,000 neurons in over 40 areas so far.
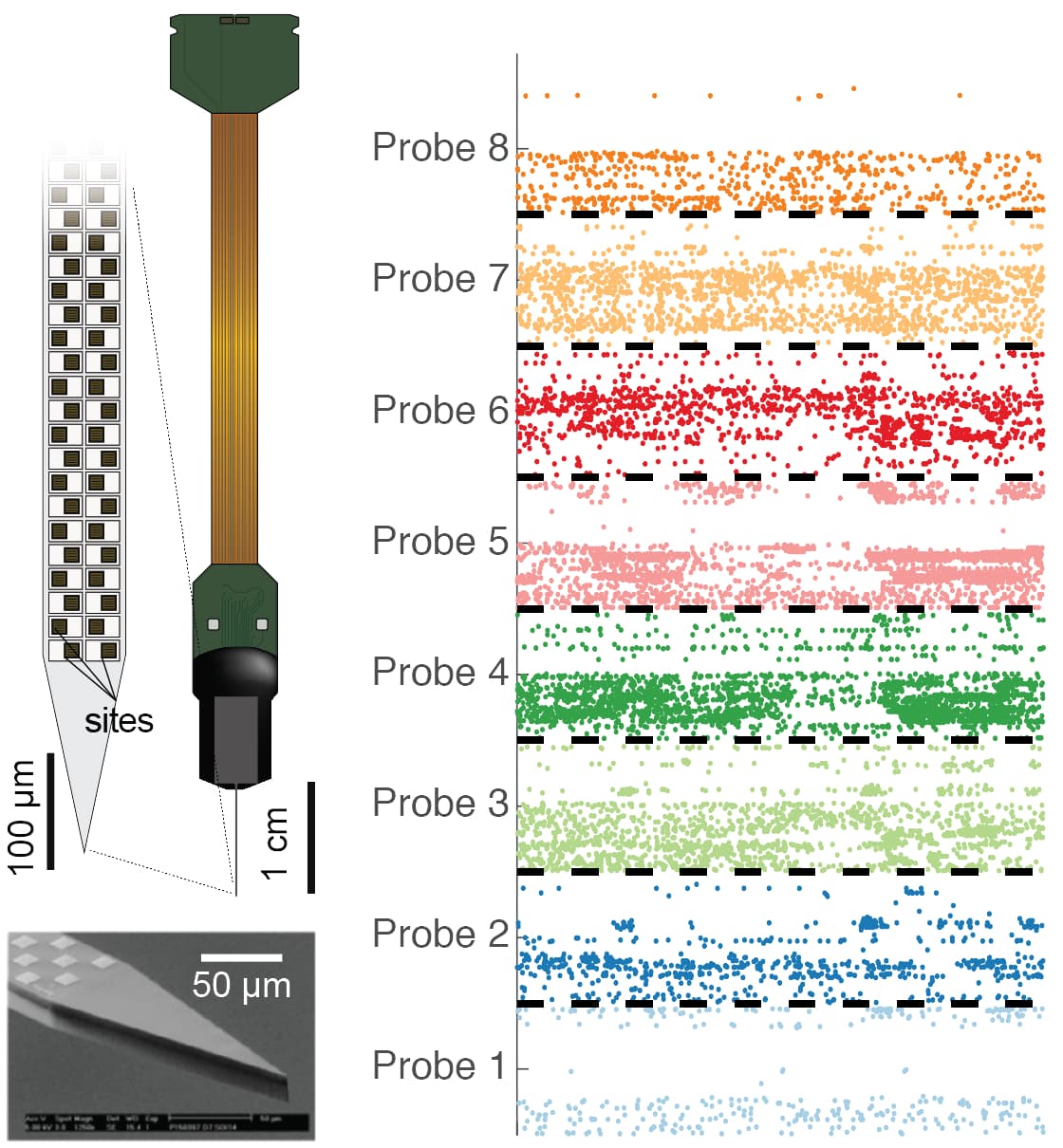
(Left) The Neuropixels probe is thinner than a hair and contains 960 recording sites. (Right) Neuron signals recorded from 8 Neuropixels probes simultaneously.
What happens in the brain during this task? Many neurons displayed strong task-related activity, meaning they actively sent signals upon the appearance of a visual cue and the motor response to it. Furthermore, there seemed to be a clear progression of task-related activity flowing through multiple key regions, starting from regions closely associated to the eye, to regions related to motor output. The entire process took place at a timescale of 100ms, validating the need of recording probes to measure these signals. Looking closer at the response of the individual cells revealed that representations of visual cues were restricted to visually-related areas as expected, while movement responses were surprisingly represented in neurons from nearly all of the recorded regions. Understanding where we find these neurons and how this kind of information flows throughout the brain is going to be key for understanding how our brain makes decisions.
Building brain atlases
Just like the 17th century explorers were navigating the earth and communicating their discoveries in colorful maps, modern neuroscientists look for ways to represent their new findings in a usable format. Brain atlases do just that. By sharing the data, brain researchers can piggyback on this wealth of information and propel their research forward. The emergence of new methods and tools gives an extra boost to the development of these extensive databases. Dan Denman, neuroscientist at the Allen Institute, discusses two brain mapping initiatives from the Allen Institute, in which Neuropixels will play an important role: the Allen Brain Observatory and the Allen Cell Types Database.
The Allen Cell Types Database is a large-scale effort to catalogue and classify brain cells from across the mouse brain. Besides the transcriptomics data, it also offers physiological profiles and morphological reconstructions of neurons. Especially for the latter two characteristics, Neuropixels created a link from the recordings of neuron signals to cell types. “The traditional way is to look at parameters of the shape of signal, the waveform, which gives a hint to the cell’s morphology,” explains Dan Denman. “It works already very well with a few or even a single parameter, for example the duration from peak to trough of the wave, but there are still limitations. We cannot always separate each cell reliably into a cell type based on a single waveform. This is where the Neuropixels probe comes in. Because the waveform of a single cell is picked up by 10 to 20 channels, we can pull out more than just a few features, such as the spread of the signal around the probe and how that spread propagates over time, including the velocity and direction.”
Even in a single brain area the extra information hints at cell morphology just by looking at the waveform. Neuropixels holds a great potential for classifying cells of many more regions with many more morphologies.
With better feature extraction, links between these recording experiments and cell type will become even stronger.
Neuropixels was also used to complement the imaging data from the Allen Brain Observatory with probe recordings of single-neuron signals. The Observatory is a database of neuron responses captured with two-photon calcium imaging in mice looking at different images. Unlike calcium imaging, probe recordings are a direct measure of brain electrical activity. “Using the Neuropixels data provides a context to the available imaging data and offers additional advantages. The ability to record from deeper structures supplements the depth-limited two-photon images very well. For example, we have less information about the thalamic inputs to the primary visual cortex. By inserting one Neuropixels probe through both regions, we can simultaneously study single-cell responses from thalamic and primary visual cortex cells. It also allows us to ask a different style of questions about dynamics and temporal properties that are mostly unavailable using calcium imaging data. For instance, we can examine response times of neurons across regions and how that response timing of individual cells relates to a broader population. Such analyses are only available to the Neuropixels probes and can help us understand the data that we have in the Observatory - and ultimately what is going on in the brain.”
The human brain on a chip
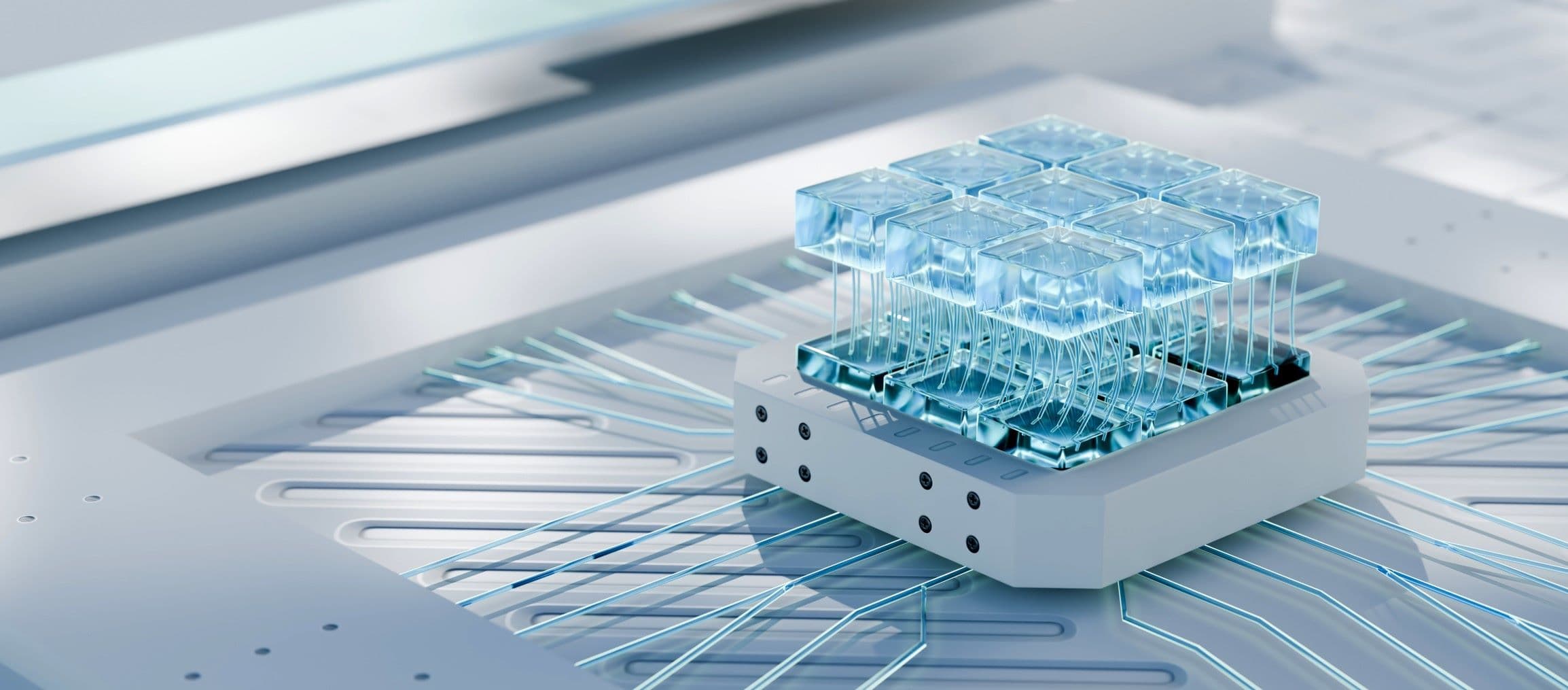
Imec’s chip technology can be leveraged to build enormously powerful instruments to study the brain, such as the Neuropixels probe, and other organs. The recent multi-electrode array (MEA) can measure signals from electrogenic cells like neurons and heart cells on a total of 16,384 sensors or electrodes. Unlike the Neuropixels probe, the imec-MEA is an in vitro device recording signals from cultured cells. However, because of a specific patterning these cells grow in a structured way on the electrodes as they would do in a real heart or brain. We are effectively growing mini-hearts and mini-brains on chip. For certain studies, this high-throughput MEA is a big step closer to mimic real organs as in human bodies.
Want to know more?
- Neuropixels probes are the next-generation recording devices for the rodent brain. They were developed through a collaboration funded by Howard Hughes Medical Institute, Wellcome Trust, Gatsby Charitable Foundation, and Allen Institute for Brain Science. The head of the collaboration is Tim Harris at HHMI Janelia Research Campus, and the electrodes are designed and fabricated by imec. The Neuropixels probes will be commercially available for researchers from July 1, 2018 onwards.
- You can find the imec press release of the Neuropixels probe here
- You can find the original Neuropixels publication in Nature here: Jun, J., Steinmetz, N., Siegle, J. et al. Fully integrated silicon probes for high-density recording of neural activity. Nature 551, 232–236 (2017). https://doi.org/10.1038/nature24636.
- Here you can find more information on the imec-MEA

Nick Steinmetz is a senior research associate in the lab of profs. Matteo Carandini and Kenneth Harris at University College London. His research at UCL focuses on understanding the neural circuits and systems that underlie perception and cognition across the brain. He approaches these questions through a combination of large-scale electrophysiology with Neuropixels probes, calcium imaging across neocortical areas, and systematic optogenetic manipulations, all in combination with a sophisticated behavioral task for mice. Before moving to London, Nick studied visual attention in the lab of prof. Tirin Moore at Stanford University.

Dan Denman is a scientist in the Neural Coding Department at the Allen Institute for Brain Science. Prior to joining the Allen Institute, he studied visual processing in the retina at Reed College and in the thalamocortical loop at the University of Pennsylvania. His current research focuses on how the brain uses populations of neurons to encode and transform visual information. To this end, he uses quantitative psychophysics and a combination of in vivo imaging and large-scale electrophysiology to record across temporal and spatial scales in the mouse visual system.
Published on:
28 June 2018