This article is the second in a series of articles focusing on how semiconductor technologies can revolutionize proteomics.
Want to stay informed about how semiconductor technologies revolutionize proteomics?
Proteins are the most relevant molecules for health monitoring – more even than DNA or RNA – because they perform the actual biological processes that make up living cells.
Unfortunately, protein analysis is much tougher than DNA/RNA analysis because these molecules cannot be easily amplified (compared to PCR (polymerase chain reactions) for DNA/RNA). Therefore, proteomics – the study of proteins – is a challenging new research field that requires systems that can identify and quantify all or specific proteins in a sample, and even of individual cells (single-cell resolution).
One primary challenge is that proteins are present in various concentrations (and sizes): from the highly abundant hemoglobin to trace proteins such as PSA (prostate-specific antigen). Protein sample preparation is becoming the technical hurdle for proteomics.
Sample preparation consists of enriching the (class of) proteins you want to identify, further purifying them, and finally separating them from non-relevant or ‘background’ molecules. Today, this is mostly performed via bulky machinery or robotic pipetting stations.
If the goal of future proteomics is to evolve towards truly compact, high-throughput systems, and to move from labs to decentralized clinical settings, it calls for an integrated sample prep.
The most popular protein sample prep systems on the market today use magnetic beads for protein enrichment, purification and separation, but this approach is hard to miniaturize or integrate and thus other routes need to be explored.
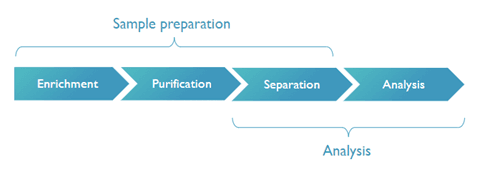
The different process steps that need to be integrated in a proteomics system, to enable truly automated, compact and high-throughput applications for use in e.g. decentralized settings such as hospitals.
Stay informed about how semiconductor technologies revolutionize proteomics
We present three technologies that can revolutionize proteomics, using active fluidics for sample preparation. They enable automation, miniaturization, integration, and multiplexing.
On-chip, high-performance liquid chromatography for protein separation
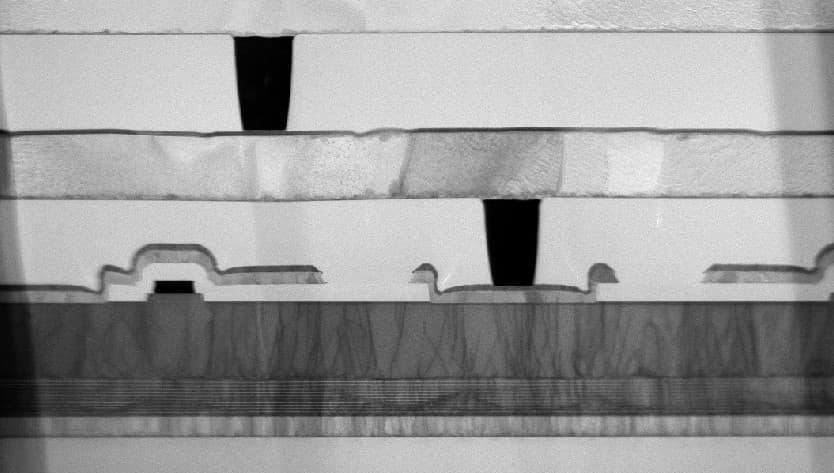
Imec designed and fabricated silicon-based high-performance liquid chromatography (HPLC) for protein separations. The technology uses silicon micropillar arrays with sub-µm to 100s µm-pillar gaps. The highly precise pillar spacing greatly improved the separation resolution compared to randomly packed microbeads in commercial systems.
In addition to the small lateral dimensions, the deep silicon etching, wafer-level quality control and the surface chemistry supported by the imec fab capability were essential to turn this technology into a successful product.
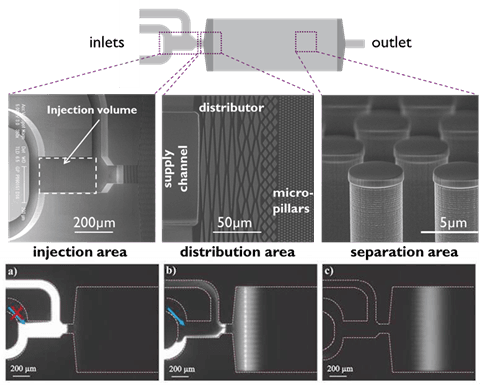
System concept and SEMs of microfluidic components for on-chip HPLC.
Integrated electrical-field gradient (EFGF)-based protein separation
Imec has strong expertise in fabricating electrode arrays with high accuracy and with dedicated materials or material stacks. Such electrodes can be used to realize electrical-field gradient focusing (EFGF) for electrically programmable, online protein separation.
Our researchers built a device concept using microfluidic channels with integrated electrodes at the top and bottom of the fluidic channels. By carefully programming and steering the electrodes, an electrical field gradient is set up longitudinally along the channel.
When a sample is introduced into the channel, the proteins experience two opposing forces: an electrostatic force – due to the electrical field and the protein’s charge –, and a hydrodynamic force – from the flow that carries them.
When the two forces balance each other, the protein molecules stop moving and form a band. As such, protein molecules are continuously enriched as the flow brings more of them, and different proteins form different bands due to their unique charge and size.
The resulting separation profile provides information about the protein composition of the sample and allows for the identification and quantification of individual proteins. Later on, the protein bands can also be released programmably.
This device concept can be further built upon, using the broad set of building blocks that imec has available for full integration into a compact system. Think for example of silicon nitride photonics, or other microfluidic components.
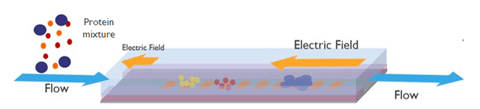
Device concept using electric-field gradient focusing (EFGF) to separate proteins. Separation is done by balancing the electrostatic force and hydrodynamic forces in suspension.
Programmable droplet processor
To overcome the diversity and complexity of different sample preps, imec developed a technology called programmable droplet processor (PDP). The device generates, moves, merges and splits microdroplets in a programmable way.
Such systems typically consist of a transistor-integrated substrate (e.g. glass), with electrodes and a thin dielectric layer on top. By selectively applying voltage to different electrodes, the local electrical field alters the charge distribution in the dielectric layer and the droplet.
Consequently, the droplet-device interfacial energy is perturbated, a force generated, and the droplet is actuated without the need for pumps or valves or even microfluidic channels. The programmable droplet processor works in a very versatile manner, so that the same hardware can be programmed and adapted to different assays. It allows for easy fluidic handling automation, signal digitization and high-volume manufacturing for multiple applications.
For proteomics, such a ‘droplet processor’ could be used to automate an enrichment, purification and separation protocol with proteins inside the droplets and precisely controlled mixing, titration, dispensing, and reaction timing events from sample prep to analysis.
Based on its expertise in transistor circuit design, electrode arrays, coating chemistry and system integration, imec designed a droplet processor technology for higher throughput, better stability, reliability and compatibility with various assays than current systems.
The PDP is integrated with driving electronics based on CMOS or TFT (thin-film transistor) technology which enables to steer millions of droplets with pixel-level of droplet occupation and behavior detection for online droplet actuation control.
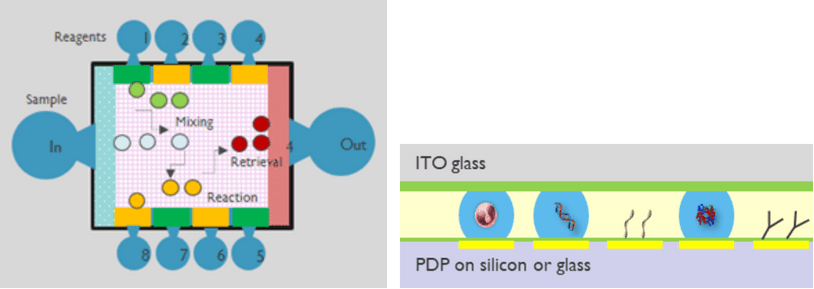
Droplet processor concept offering a reliable and high-throughput solution for integrated sample prep and analysis assays in proteomics.
In conclusion, proteomics presents significant challenges and there is a growing need for integrated and compact systems, especially for decentralized settings like hospitals. Three promising approaches for integrated sample prep have been presented. These approaches offer automation, miniaturization, integration, and multiplexing capabilities.
Together with interested parties, they can be further developed and integrated into a company’s specific product and protein assay, significantly impacting the field of proteomics and enabling new insights into protein analysis and function.

Dr. Chengxun Liu is currently a program manager and principal member of technical staff in the life science department at imec. His main responsibility is to identify and develop microfluidics solutions for life science applications. Chengxun has a mixed background of engineering and biology, with the belief that microtechnology enables innovative solutions to life science challenges. His research interest and experience covers a broad range of biophysics and microfluidic technologies, such as cell isolation, molecular assays, electrokinetics and droplet fluidics. Being a principal scientistn Chengxun has been leading the development of several technologies such as single-cell electrical impedance spectroscopy, high-speed cell sorting, droplet sorting & programmable processor, electrical cell & biomolecule manipulations, etc.
Published on:
3 May 2024