February 29 is Rare Disease Day. To mark this occasion, imec researcher Roel Wuyts shares his vision on how personalized and mobile sequencing will accelerate the diagnosis of - and research into - rare diseases.
The odyssey that comes with rare diseases
More than 300 million people around the world suffer one or more of the approximately 6,000 identified rare diseases. Because only a handful of people suffer the same disease, diagnosing and treating these diseases is difficult. The whole process starts with a doctor but builds up to a veritable odyssey, with patients being sent from one doctor to another.
For the doctor too it is difficult to come up with the right diagnosis and start adequate treatment. Currently what often happens is that during an afternoon slot at a conference, a doctor gives a short presentation about his/her patient’s symptoms. Sometimes there is another doctor in the room who recognizes these symptoms from one of his/her patients. This is how a rare disease is discovered: it remains a highly primitive process and the chance of success is limited.
The GAP project bridges this gap
To improve that process, researchers at KU Leuven developed specialized software and then joined forces with imec's ExaScience Life Lab to further develop the software and the underlying platform. This took place as part of the imec.icon project ‘GAP’ (Genome Analytics Platform).
According to Roel Wuyts, team lead at the ExaScience Life Lab, personalized and mobile sequencing will provide a huge impetus for the diagnosis process. To mark Rare Disease Day, he communicates his vision on how this technique can be used in the fight against rare diseases.
Approximately 70% of rare diseases result from a genetic disorder. As part of the GAP project, KU Leuven and the ExaScience Life Lab have developed a software platform in which doctors can upload their patients’ genetic code and phenotype.
“Doctors in participating hospitals can use the WiNGS software to search for similar external characteristics and indications in the DNA. If there are positive results, the doctors of both patients can come into contact with each other, just as they would at a conference, but then online rather than in person. Furthermore, the privacy of the search query is guaranteed, so that the patient’s data, and that of the doctor using the search function, are protected,” says Roel Wuyts.
DNA as the basis for the diagnosis process
The system evidently requires that the DNA is already available, ideally with some interesting variants. For this purpose, first a trio analysis is conducted, where you compare the patient’s DNA with that of both biological parents. In the majority of cases, the subject is a baby, given that 70% of rare diseases manifest themselves at a very young age. If the parents do not have the disease, the baby’s DNA can be searched to find anomalies - often in a combination of genes. Of course, variants will also be discovered that have nothing to do with the disease, because every person is unique, but in this way the list of candidate genes is already significantly reduced and the search consequently made more efficient.
However, we still want a quicker way to incorporate sequencing and DNA in the diagnosis process. A lot of work has gone into this already in GAP, and from a software point of view, things have already been speeded up. But sequencing requires more than that. Sample preparation takes a day’s work and afterwards another couple of days are required for the sequencing itself. When you include the software aspect, you have quickly spent 3.5 days.
“If you want to better support the diagnosis of rare diseases through, among other things, the use of trio analyses, the process needs to get substantially faster, simpler and cheaper. This is why, as part of personalized medicine, we want to push forward personalized mobile sequencing as a solution,” says Roel Wuyts.
Personalized mobile sequencing: from sample to DNA in 4 hours
For personalized and mobile sequencing, you need two ingredients: a device, and population genomics. The device must allow you to go from a raw sample to an analyzed result in 4 hours (instead of 3.5 days). Because the whole process - from sample to result - occurs in the same device, it also runs much more efficiently.
“For rare diseases, personalized and mobile sequencing is a game changer. Imagine that the doctor can’t immediately tell what is wrong with your baby, you will in principle be able to upload a sample on the maternity ward and 4 hours later find out if it is a known rare disease. In this way you are not dependent on the doctor’s knowledge of all rare diseases, and the odyssey has immediately become much shorter”.
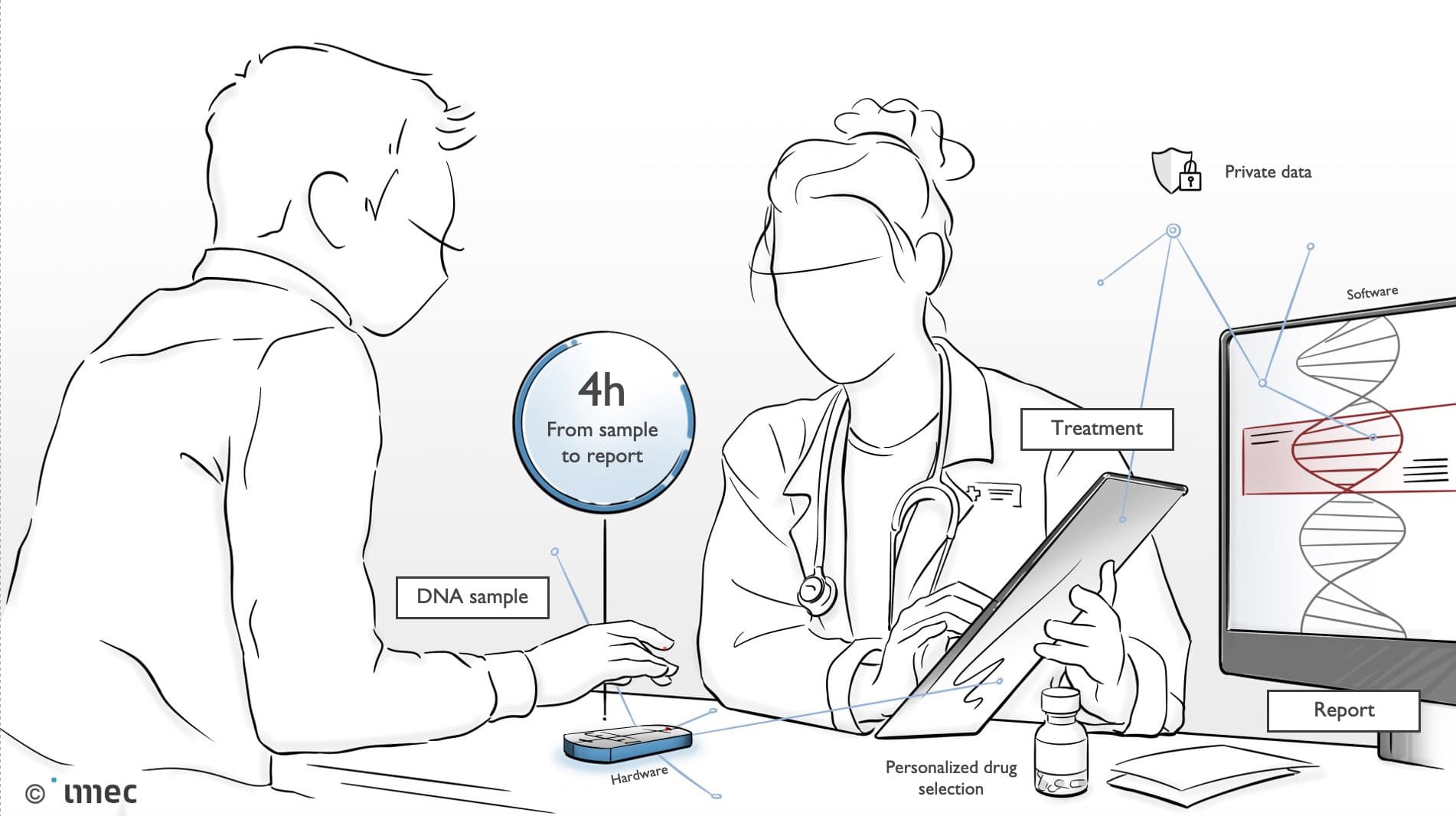
Personalized mobile sequencing brings the sequencing time down to 4 hours.
“But to be able to make such diagnoses, you also need DNA…a lot of it. In rare diseases you have to look at the whole genome because this is not about one disease but a collection of diseases. Additionally, apart from the DNA, you need a lot of information about all these different rare diseases because before you can say that there is a case of a rare disease, a lot of analyses must have already been done. This is what we call large-scale population genomics”.
A solution in the near future
“The next couple of years, imec wishes to cover both aspects: in general for personalized medicine, but also specifically for rare diseases. We already have a lot of the technological building blocks to achieve the 4 hours, e.g. our work on microfluidics, the actual sequencing chips, the specific software, and the work on neuromorphic computing to make the analyses quicker. However, work is still required to integrate all the elements. Realistically, I think that we can achieve a personalized and mobile sequencing device in 4 to 5 years,” explains Roel Wuyts.
Privacy protection will be a crucial component. “In 4 to 5 years, a whole lot more genomic and clinical data will exist, but it will also be spread out. In this data, we want to look at large-scale population genomics in a way that is privacy-protected so that the data does not need to be aggregated. That is however difficult from a practical point of view, because you would need a very large database for all the genome data. Plus, where you would put such a database? For this project, we are thinking on a European scale and beyond, but some countries for example have rules on medical data not leaving the country. We are therefore turning to techniques that protect privacy, an area in which ExaScience has years of experience, so that all sequencing devices can be connected to the same system”.
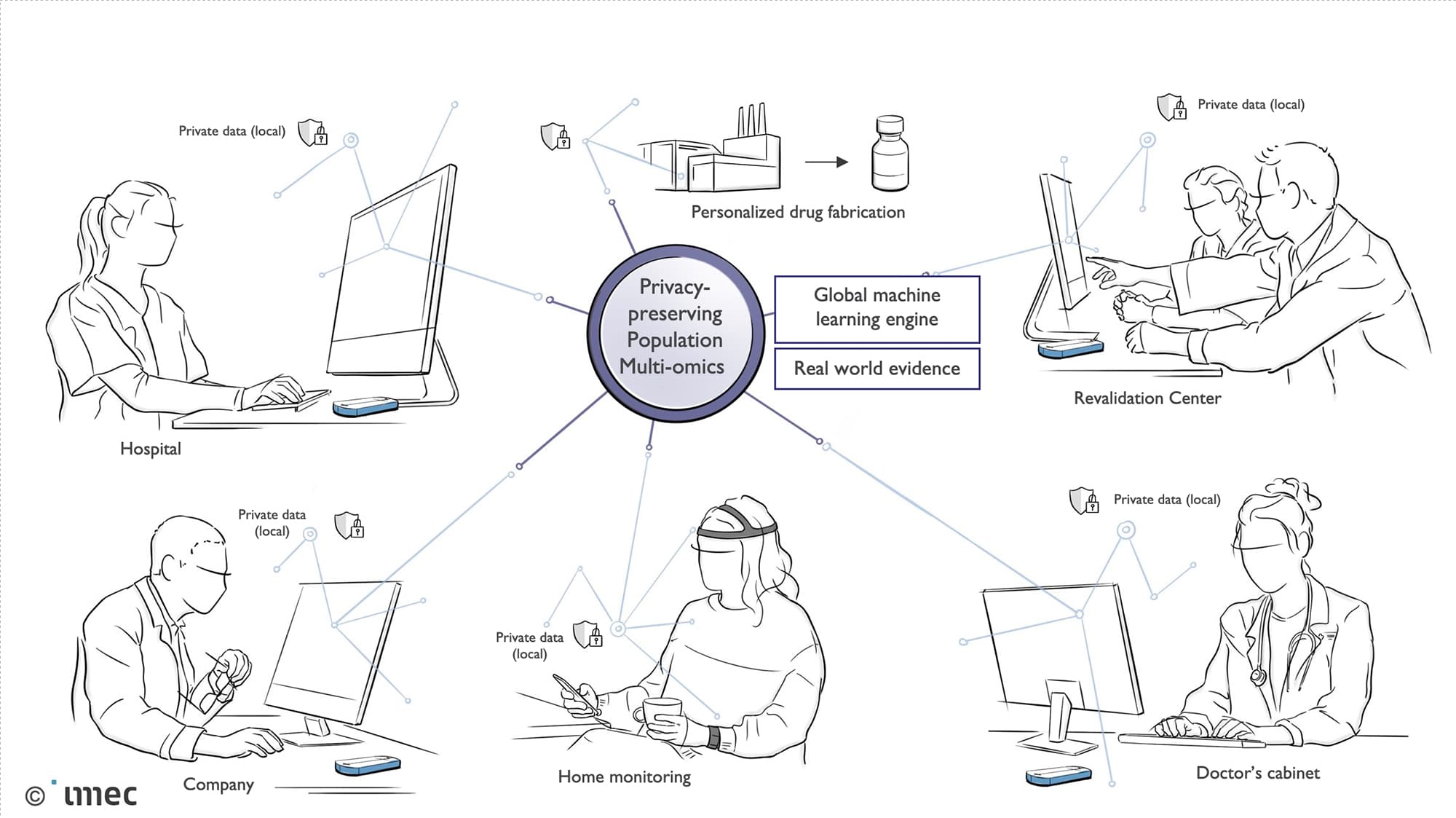
Privacy-protected techniques for population genomics.
What’s in it for rare diseases?
A personalized mobile sequencing device is appealing for different parties. Firstly, doctors and their patients gets answers more quickly, without having to embark on an odyssey. Secondly, researchers can use the system to find new anomalies. They can, for example, apply machine learning to the data to find correlations. As such, unknown rare diseases can be searched for proactively. Finally, it is also handy for the pharmaceutical industry, which will be able to look for potential targets among new and old rare diseases in order to develop medicines. The fact that such large-scale data collection can be approached in a privacy-protected way makes it very interesting for this target group.
Would you like to find out more?
- More information about the imec.icon GAP project can be found on their website.

Roel Wuyts is heading imec’s ExaScience Life Lab, which focuses on upscaling software solutions for data-intensive high-performance computing, primarily (though not exclusively) in the domain of life sciences.
The ExaScience Life Lab has many years of experience with high-performance computing technologies, programming languages and the use of hardware accelerators. We often help companies develop prototype solutions for complex problems involving several disciplines. We have successfully done this in the past for large-scale projects on the subject of machine learning (for pharmaceutical companies and imec projects), DNA sequencing software for hospitals and pharmaceutical companies, advanced biostatistics and data analysis, and even Industry 4.0 and space weather simulation.
Published on:
3 March 2020